Hi!
I’m currently using the WheatExpression Browser (precisely, the BLAST Scaffold feature at the right side of the browser) to fish-out the wheat ortholog(s) of a rice gene; I’m facing a couple of issues:
Issue#1: Error: Something went wonky
Looks like you have encountered a bug in SequenceServer. Could you please report this incident to your admin or to SequenceServer Google Group (if you are the admin), quoting the error message below?
The programme used was ‘tBLASTn’ and this is only with the case of the database ‘TGACv1’; the error message is attached.
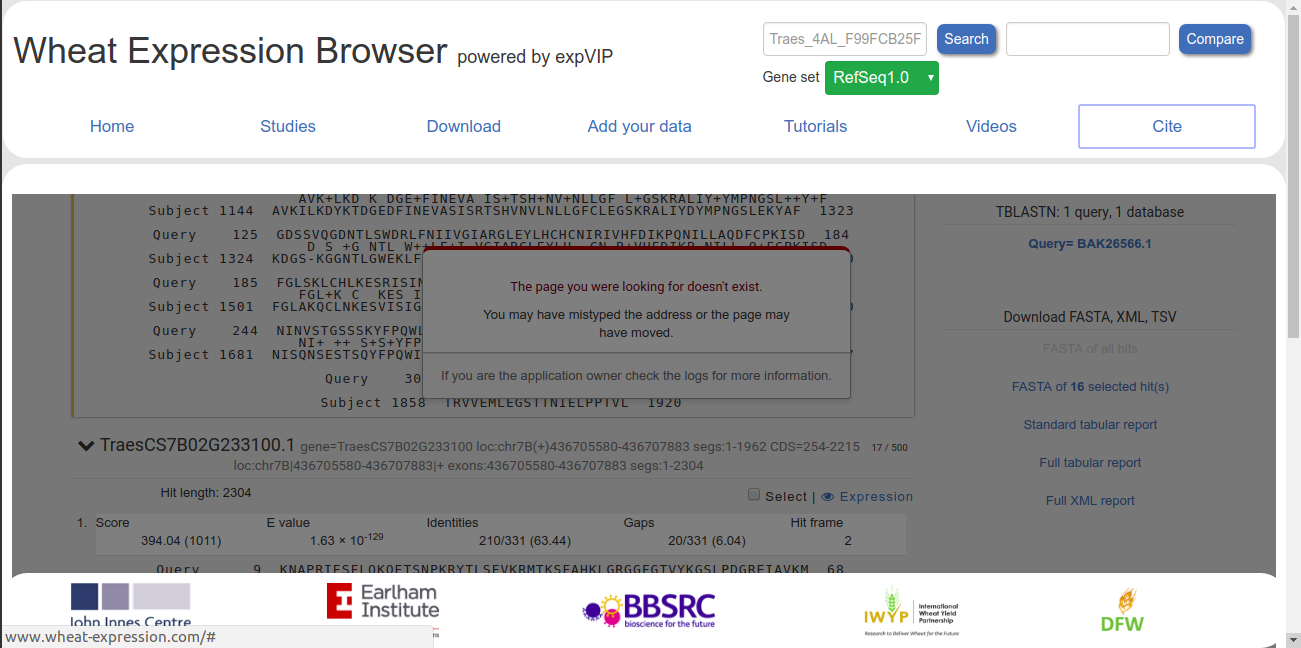
Issue#2: When I select the download options (the fasta of the selected hit(s), full tabular report, etc) at the right-hand side of the browser, I’m shown an error message which is also attached.
The .fa file which I used as query also is attached.
Please help me download my desired sequences.
Thank You,
Hari G,
PhD Scholar,
NIPGR, Delhi
Issue#1 (8.96 KB)
Hari_Sequence.fa (381 Bytes)