Hi,
I am a novice to using Linux. I got sequenceserver to run in ubuntu 14.04.
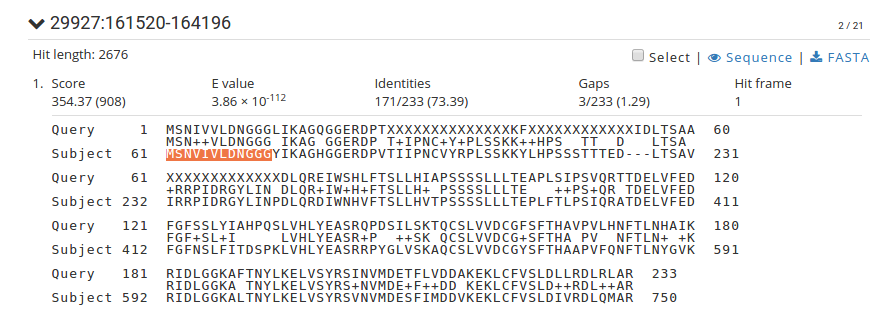
I had performed tblastn and was going through the results. The coordinates of the subject in the alignment lines were not in concordance to the alignment. Please find the screenshot of the alignment (sequenceserverlocalblast.png) in the attachments with this. Please go through the start- and end-amino acid residue-coordinate of query and subject in each alignment line in the attached files.
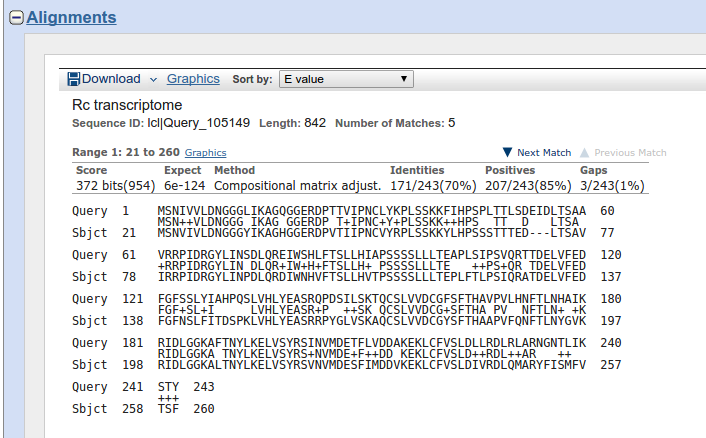
To ensure the problem, I used “align two sequences (protein)” option of ncbi-blast to align my protein query against the corresponding translated sequence (subject) from the local database. I got the same alignment pattern as from using sequenceserver, except that the alignment coordinates were fine (attachment: NCBI_proteinBlast_align2seq.png) in the later case. Is this a bug in the program or have I not accounted for some error on my part? Please clarify.
Thanks