Hello, friends!
Can you tell me how to install a database server in the sequenceserver for blast?
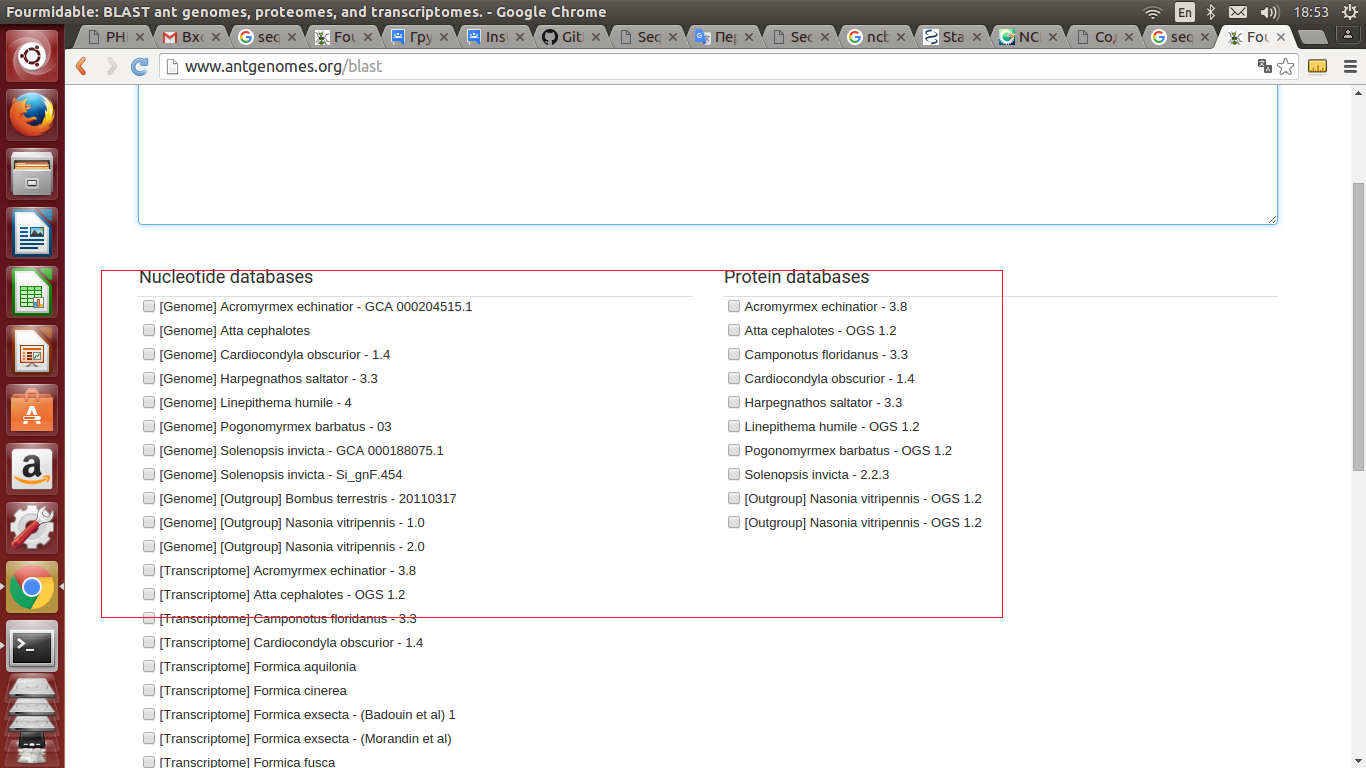
I need to bind the database for the blasts as in your screenshot 2_2.png.
But I don’t know where to download the file on site http://www.ncbi.nlm.nih.gov/, for example, such as viruses and install the database.
I see http://www.ncbi.nlm.nih.gov/books/NBK52640/ and see database ftp://ftp.ncbi.nlm.nih.gov/blast/db/ and I don’t install the database for sequenceserver.
Thank you very much!